Cell-cell communication mediated by secreted proteins from scRNA-Seq data
Source:vignettes/scCCC.Rmd
scCCC.RmdThis tutorial demonstrates how to infer secreted protein-mediated cell-cell communication (CCC) from scRNA-seq data. This analysis requires an scRNA-seq dataset with two distinct conditions. SecAct employs two key criteria to identify condition-specific cell-cell communication mediated by a particular secreted protein: (1) upregulated gene expression of the secreted protein in one cell type (defined the source cell type), and (2) increased signaling activity of the same secreted protein in another cell type (defined the receiver cell type). For this demonstration, we employ an ovarian cancer (OV) scRNA-Seq study containing samples from both primary and metastatic sites.
Prepare expression matrix
The OV scRNA-Seq data is stored in a Seurat object. The information
of cell-type annotations and anatomical sites are stored in the columns
MyCellType and Groups of
meta.data from this Seurat object, respectively. Users can
easily access them using the following code.
library(SecAct)
library(Seurat)
# download data from https://hpc.nih.gov/~Jiang_Lab/SecAct_Package/OV_scRNAseq_Seurat.rds
# load scRNA-Seq data
Seurat_obj <- readRDS("OV_scRNAseq_Seurat.rds")
# cell count
table(Seurat_obj@meta.data[,"Groups"])
## Metastatic Primary
## 18746 17589
# show UMAP plot
my_cols <- c(
'B'='#C88888','CD4T'='#3288bd','CD8T'='#B95FBB','DC'='#63636d',
'Endothelial'='#D4D915','Fibroblast'='#66c2a5','Macrophage'='#ff9a36',
'NK'='#bb8761','Tex'='#fe666d','Treg'='#E6C122','Tumor'='#A4DFF2'
)
p1 <- DimPlot(Seurat_obj, reduction = "umap", cols = my_cols, group.by = "MyCellType")
p2 <- DimPlot(Seurat_obj, reduction = "umap", group.by = "Groups")
p1+p2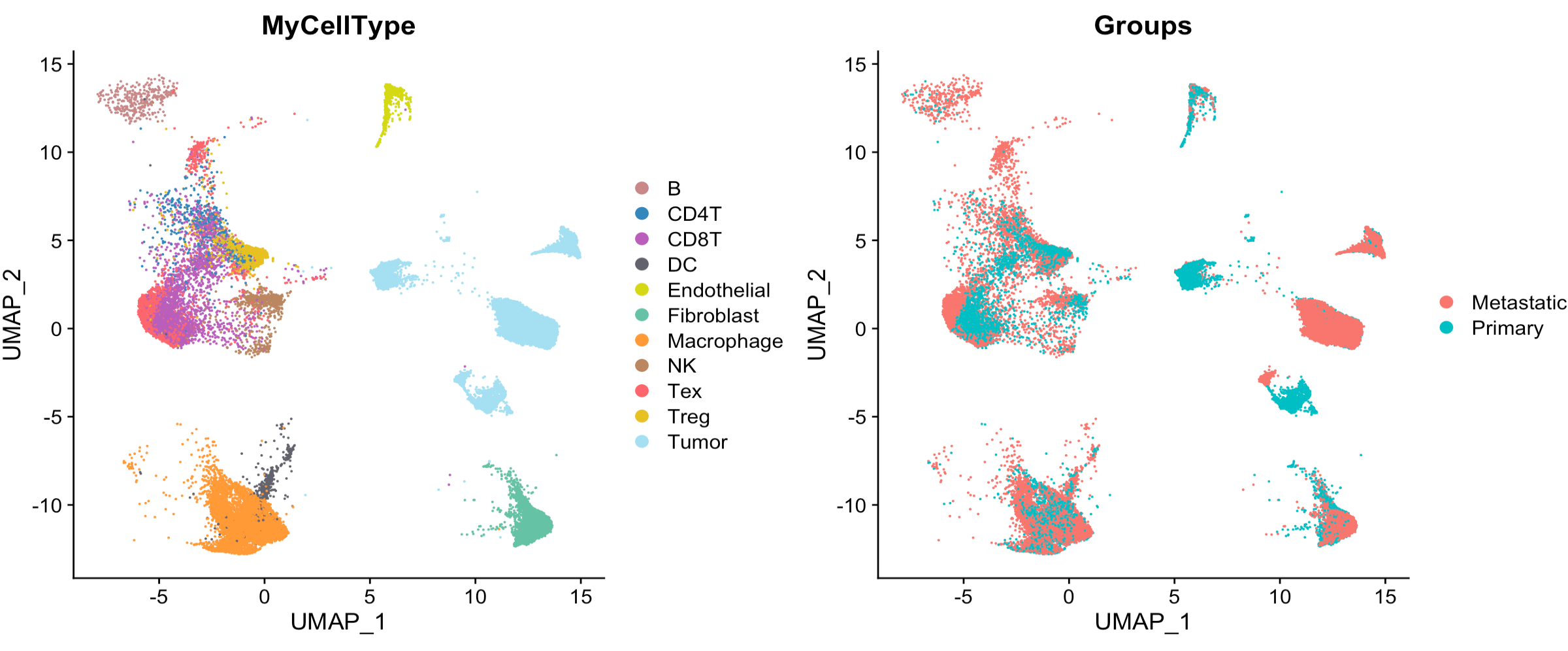
Infer cell-cell communication
SecAct performs three steps to infer condition-specific
cell-cell communication mediated by secreted proteins: (1) assessing
changes in secreted protein expression for each cell type (defined the
source cell type) between two conditions (e.g., Metastatic vs Primary),
(2) calculating changes in secreted protein signaling activity for each
cell type (defined the receiver cell type) between two conditions, and
(3) linking source and receiver cell types.
When running SecAct.CCC.scRNAseq, user need to assign
the cellType_meta and condition_meta with the
column names of cell-type annotation and condition information in
Seurat_obj@meta.data, respectively. Also, claim what case
and control conditions are by conditionCase and
conditionControl. User can find the SecAct results in
Seurat_obj @misc $SecAct_output.
# infer CCC; ~2 mins
Seurat_obj <- SecAct.CCC.scRNAseq(
Seurat_obj,
cellType_meta="MyCellType",
condition_meta="Groups",
conditionCase="Metastatic",
conditionControl="Primary"
)
# show output
names(Seurat_obj @misc $SecAct_output)
## [1] "SecretedProteinExpression"
## [2] "SecretedProteinActivity"
## [3] "SecretedProteinCCC"
# cell-cell communication results
head(Seurat_obj @misc $SecAct_output $SecretedProteinCCC)This table lists metastasis-specific cell–cell communication events mediated by secreted proteins. Each row represents one interaction, indicating that the corresponding secreted protein shows upregulated expression in the sender cell type and increased signaling activity in the receiver cell type under metastatic compared with primary conditions.
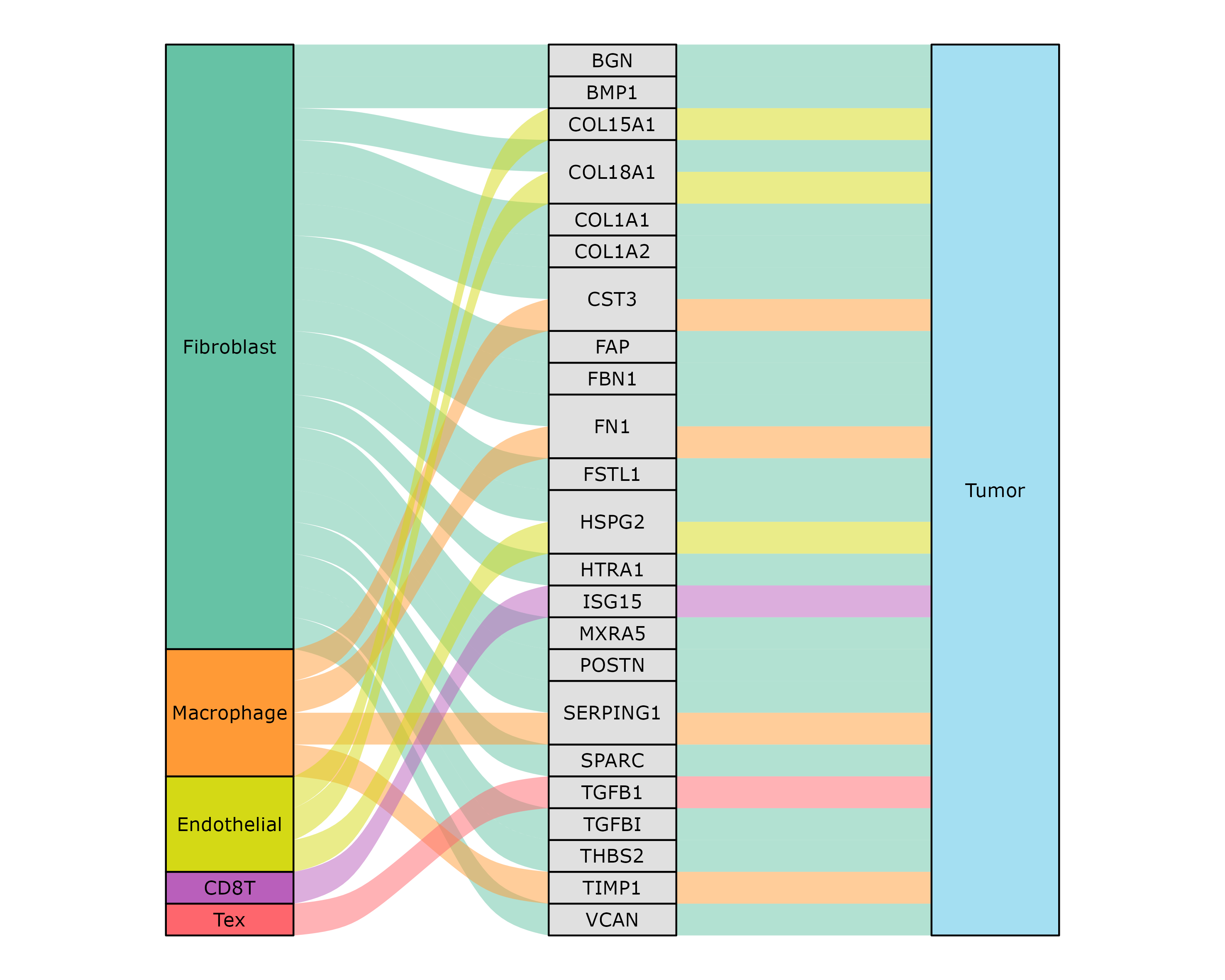
Visualize cell-cell communication
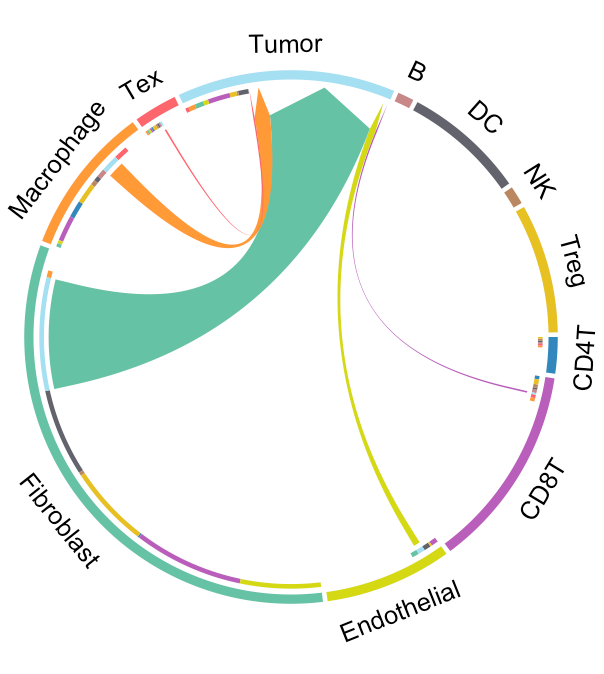
We provide two types of visualization plot for cell-cell communication, i.e., heatmap and circle plot. The number in the heatmap represents the count of secreted proteins from senders to receivers.
SecAct.CCC.heatmap(Seurat_obj, row.sorted=TRUE, column.sorted=TRUE, colors_cellType=my_cols)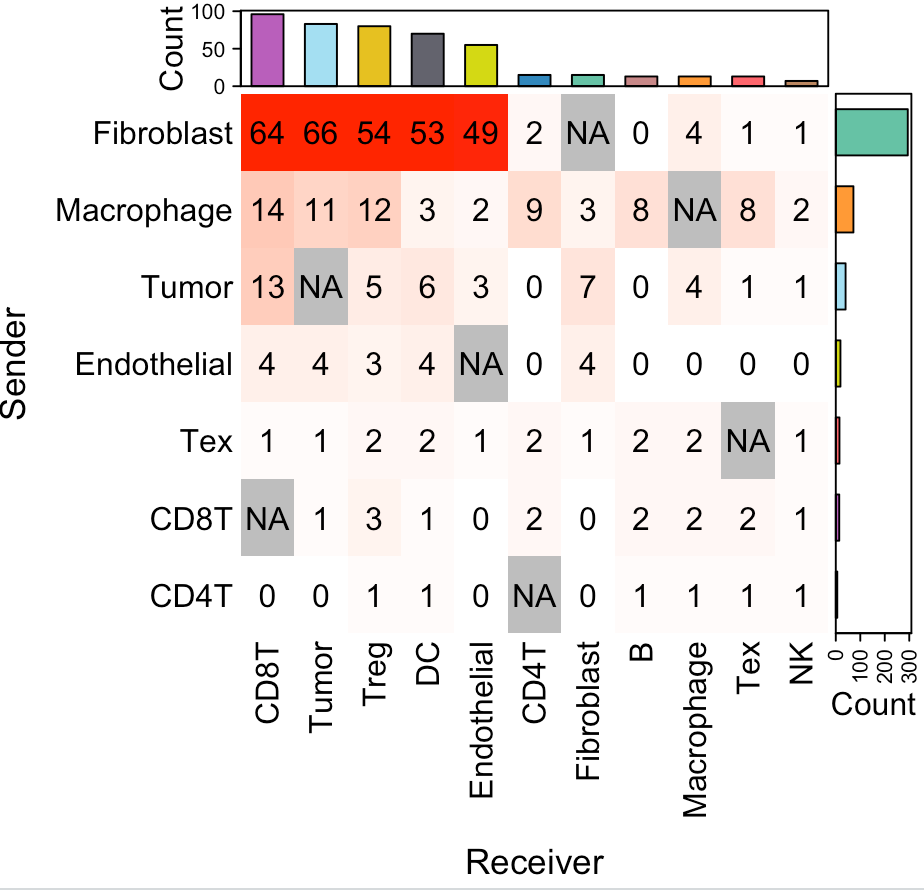
SecAct.CCC.circle(Seurat_obj, colors_cellType=my_cols)
SecAct.CCC.circle(Seurat_obj, receiver="Tumor", colors_cellType=my_cols)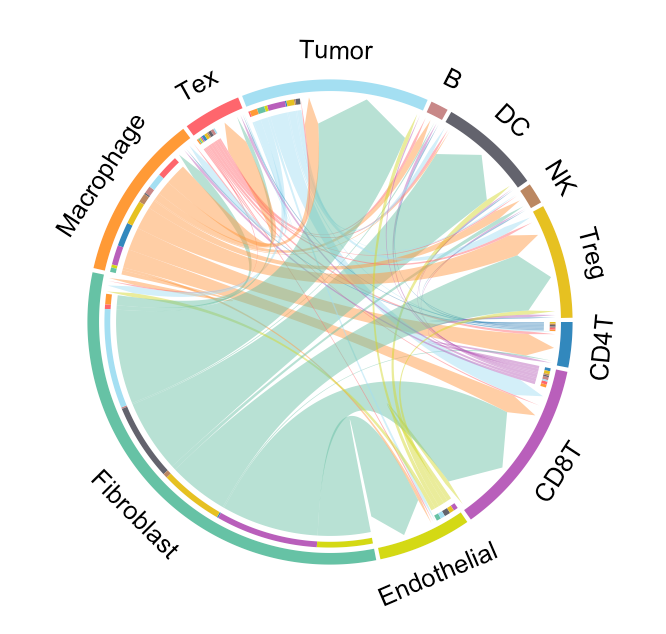
Visualize secreted signaling flow
User can select the cell-cell communication of interest from
Seurat_obj @misc $SecAct_output $SecretedProteinCCC to
visualize them using SecAct.CCC.sankey. You need to assign
values to sender, secretedProtein, and
receiver.
sender <- c("Fibroblast","Macrophage","Endothelial","CD8T","Tex")
secretedProtein <- c(
"BGN","BMP1","COL15A1","COL18A1","COL1A1",
"COL1A2","CST3","FAP","FBN1","FN1",
"FSTL1","HSPG2","HTRA1","ISG15","MXRA5",
"POSTN","SERPING1","SPARC","TGFB1","TGFBI",
"THBS2","TIMP1","VCAN"
)
receiver <- c("Tumor")
SecAct.CCC.sankey(
Seurat_obj,
colors_cellType = my_cols,
sender = sender,
secretedProtein = secretedProtein,
receiver = receiver
)